
How Bota built its Synthetic Biology AI search engine
Background
With the help of “Bota Biofoundry”, our in-house developed Biomanufacturing platform that expedites our work throughout the entire DBTL cycle, the bottleneck of the speed of our research is no longer the time it takes to plan, prepare and execute the experiments. Rather, it is how fast we can generate new ideas that becomes the new limiting factor. Since the explosion of LLM (Large Language Model) based AI technologies such as the ChatGPT, we have been exploring and experimenting ways to help our scientists to generate new research ideas faster with the help of AI.
After thorough investigation, we categorized existing novel AI-powered research tools on the market into two categories:
- Global literature search tools (e.g. Scholarly AI), for finding the most relevant academic and scientific literature for user’s research needs
- Intensive paper reading tools (e.g. ChatPDF, Txyz.AI), for ChatGPT-like Q&A with the content of a single or a few PDF files
While new products in both categories has been emerging every day especially after the launch of OpenAI’s GPT-3 API, and does have some intersections in their tech stacks, it quickly became clear to us that they served vastly different purposes in the research process. Before we dived deep into this rabbit hole, we worked alongside with our scientists to understand their research needs.
As it turned out, while those ‘ChatPDF’ like products are certainly impressive in their capability of quickly extracting key information from papers, the accuracy is far from perfect and may cause the reader to omit important details from the paper. As a result, most scientists would rather read through each paper by themselves.
Instead, we believe a good global search tool (like the Google Scholar, but enhanced by AI) would truly improve the efficiency and experience of our research and idea generation processes, since the true bottleneck lies in finding the most relevant materials, and filtering out as much noise as possible from the vast ocean of information we have today.
The Problem
While Large Language Models like the ChatGPT have proven to be helpful for all sorts of use cases, including scientific research, they all suffer from a phenomenon called hallucination, where the AI chatbot creates outputs that are inaccurate, or simply nonsensical. In case of using LLMs for scientific research, they tend to hallucinate non-existing citations to papers and articles, meaning that while it may seem to save time using ChatGPT to help you find scientific literatures, you still have to do your own research and make sure that the citations provided by the AI actually exist. Unfortunately, hallucinated content can often look convencing even to the most careful eyes. Obviously, this defeats the whole purpose of using ChatGPT to improve work efficiency.
Since finding and reading scientific literature are crucial to our scientists’ research, especially for the idea generation and feasibility study stages, the Computation team of Bota set out on a journey to create our own Literature AI, aiming at the specific field of Biology that the company is focusing on: Synthetic Biology.
Our Solution
We have created an AI-powered search engine that allows users to use natural language queries to search over 130,000 PubMed papers related to Synthetic Biology and Metabolic Engineering. Like the ChatGPT, you get an instant response to your question, including the most relevant papers or articles you should read and some general suggestions on approaching your research question.
But unlike the ChatGPT, or any other LLM chatbots in general, our Literature AI does not hallucinate. Each time after you press the search button, the system actually goes through the entire database of all of the latest SynBio related papers from PubMed, determines the top 100 papers that may be useful to your question, and finally utilizes the power of the latest LLM to generate a detailed answer to your question. As a result, you can be confident that the citations in the AI answer are actually real and can be easily found in the links provided.
As a bonus, you can even “chat” with the AI regarding any questions you have about any of the papers or articles in the search results. Want to extract all the genetic modifications the authors did to their best-yielding strain? Bota Literature AI got you covered, right on the same interface!
How does it work?
PubMed is a free database containing over 37 million citations for biomedical literature, life science journals and online books. It is, inarguably, the most popular bibliographic database for researchers in the health and medical sciences.
Our plan was to create a local copy of metadatas of all the articles that may be useful to us from PubMed in our own database, and periodically update the database so that our colleagues can always find the latest studies in their search results. And we were not just creating a simple copy of the database — we want to create a vector database that contains the embeddings, aka the numerical representations of the abstract section of each article, so that our Literature AI can utilize this database to look for the most relevant articles for a user’s query based on semantic similarity.
Unlike traditional search methods that rely on keyword matching, a vector database stores data as high-dimensional vectors, which are numerical representations of features or attributes, and achieves accurate similarity searches based on vector distance or similarity. In case of literature search, we can convert the abstract sections of each article from PubMed to such vectors using an Embedding Model, and store them in the vector database. Since an abstract section usually contains the central ideas and key points to the article, and are publically available even for those paywalled articles, it is the perfect choice for performing semantic searches for our Literature AI.
Challenge 1: Creating the database
PubMed generously provides several methods for users to programmatically download all the citations from its database. At first, we tried to download them through their FTP servers (https://ftp.ncbi.nlm.nih.gov/pubmed/baseline/). The FTP site contains over 1200 XML files, each containing over 20,000 entries including metadatas such as the title, abstract, authors, etc. By going through these XML files, we were able to store the information about all 37 million papers and articles from PubMed in our own database. The calculation process of generating vector embeddings for all of these abstracts did took a while — a little less than a week for our Nvidia Tesla T4 GPU while running 24 hours a day. Not too bad, right?
With over 37 million embeddings in the database, each having 768 dimensions, the time it took to perform just a single search becomes unbearably slow. While creating a vector index (such as the IVFFlat or HNSW index) reduces the search time to seconds, due to the sheer size of the database, the computational power it requires to incrementally rebuild the index as we add newly published articles to the database becomes expensive. Instead of throwing budget blindly into this project and upgrade to a more expensive cloud server plan, we decided to try another approach instead.
To reduce the number of entries in our vector database, we need to find a way to effectively filter out any irrelevant articles to our company, while keeping all the relevant ones included so that our scientists would not miss any potentially useful information. Unfortunately, the XML files on the PubMed FTP server does not contain any useful attributes that would allow us to find all SynBio related articles from all 37 million citations.
After studying PubMed’s own search engine, we found a neat feature called the ‘MeSH Heading’ (Medical Subject Headings). MeSH is the NLM controlled vocabulary thesaurus used for indexing PubMed citations, and the MeSH terms include Subheadings, Publication Types, Supplementary Concepts and Pharmacological Actions. Among all the MeSH terms, we found two that are perfect for our use case: “Synthetic Biology” and “Metabolic Engineering”.
The XML files on the FTP does not contain any MeSH attributes. However, we were able to use the ‘Entrez’ Python library to search for articles on PubMed that contain the MeSH headings we want.
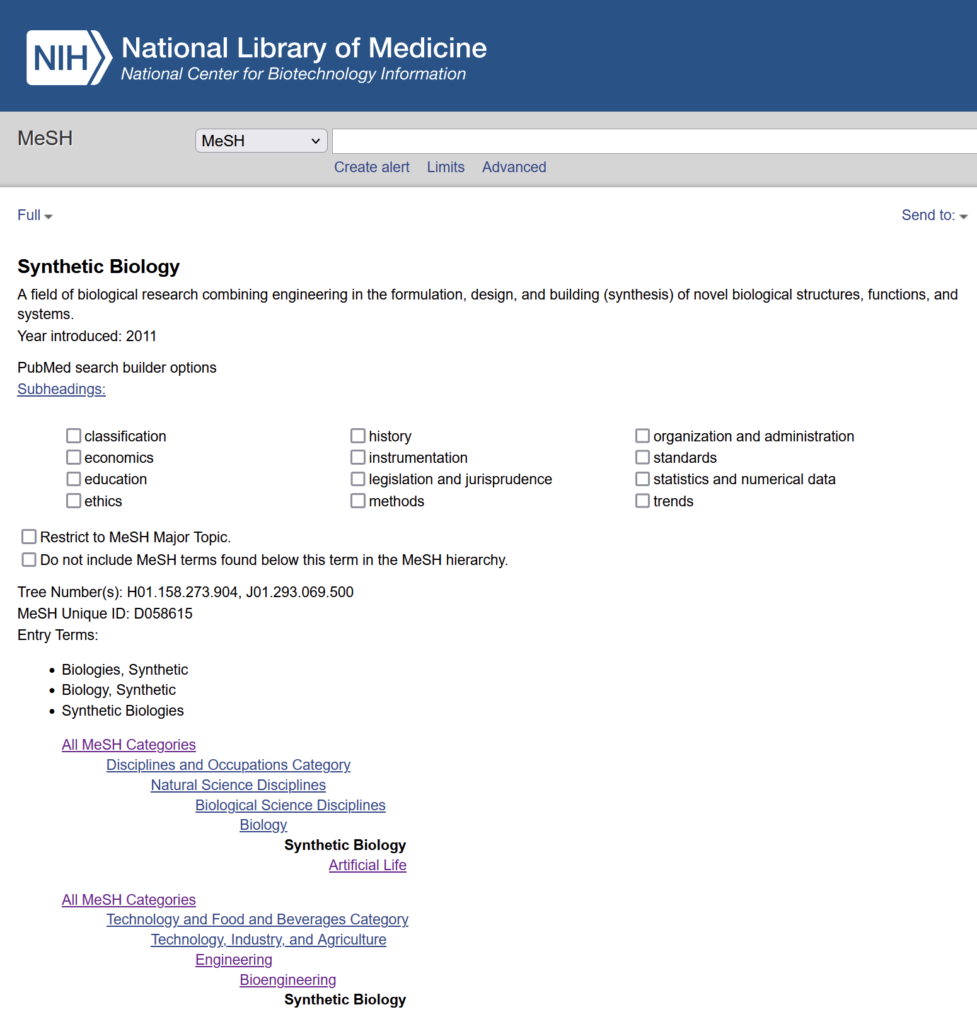
One small issue we met while trying to download citations of ALL articles that contain the MeSH terms “Synthetic Biology” or “Metabolic Engineering” is that the Entrez API will only return the first 10,000 entries for each search. Clearly, there are more than 10,000 articles related to these two subjects!
To circumvent this issue, we created a script that limits the year range of each search to 1 year at a time, and combined the search results from 1970 all the way to 2024. With this one simple trick, we were able to get over 130,000 citations of all SynBio-related articles from PubMed into our vector database. The smaller database, unsurprisingly, turned out to be much easier to handle for even the cheaper cloud database plan on Azure Cloud, allowing near-instant search results even without any vector indices.
Challenge 2: Finding that needle
Once we got the vector database going, we quickly realized that the accuracy of the search result with just the vector search alone is not going to cut it. While the top 100 results did contain articles we are looking for most of the time, they are usually not placed in the top positions of the list. Since most large language models like the GPT-3.5 have limited context length (even the newer GPT-3.5-Turbo-16k), we cannot simply throw the entire search result to the model.
To further improve the sorting of the search result from the vector database, and hence improve the response accuracy of our Literature AI system, we re-ordered the search result. To do that, we used another type of model called the rerankers.
But how are rerankers going to help if we have already used embedding models to create the vector based retriever? Unlike embedding models, which are run at the building stage of a vector database, hence having no context of user’s query, a reranker model is run at the query time. The reranker is a type of model that takes a pair of text — in this case, the user’s query and a single article abstract from the search result, and generates a similarity score. By running the reranker model against the user query and every single article abstract from our vector search result, we can use the outputted scores to effectively re-order the search result and filter out the less relevant result entries.
In addition to reranker models, another advancement in Large Language Models in 2024 has also supercharged the performance of our Literature AI significantly. On March 19, 2024, Anthropic released their latest family of large language models, Claude 3. Claude 3 Haiku, in particular, caught our attention immediately. The model promises near-instant responsiveness, costing less than the already affordable GPT-3.5-Turbo, while providing a whopping 200k context length. Of course, we were skeptical at Anthropic’s claim at first. It is one thing to support a ridiculously large input size, but another to be able to actually utilize the entire context. We were glad that our skepticism was proven to be wrong.
We built a pipeline that first gets the top 200 search results from our vector database, then downsizes the list to 100 with the help of a reranking model, and finally dumps these 100 article abstracts to Claude 3 Haiku along side with user’s query. Even though we are pushing the tiny Haiku to its 200k context limit with each and every single user query, the result has been amazing. We tried asking our Literature AI the problem statements of our existing projects, and the system was able to accurately recommend all the papers and articles our lead scientists have been referring to for those projects every single time. Not only that, Claude 3 Haiku was able to provide detailed explainations of why each article was picked among other articles in the search result, and how it may help us with our research. The model also provides link to each recommended article, giving users a peace of mind that none of the output from our system is AI hallucination.
Summary
In the end, we were able to build an AI powered SynBio literature search engine that saves hours of research for our colleagues while costing only $0.01 per search, providing not only friendly and helpful to user’s natural language queries, but also recommendations to the most relevant papers and articles, even when traditional keyword based searches failed to do so.
We are also aware of an interesting open source project named the “GPT Researcher”. While also aiming towards finding the right online resources for users’ research needs, the project uses a slightly different approach by using multiple AI ‘agents’ to conduct research from various online sources with search APIs and web scraping tools. In practice, we found that the output from this tool is still susceptible to AI hallucination, often generating convincing responses from non-existing citations. Nevertheless, we are still amazed by this lesser-known project, and will consider trying to contribute to the project, or even incorporating their work into our SynBio Literature AI.
Through this little adventure of implementing emerging AI technologies into our everyday work, we gained valuable experience in building highly customized AI-native applications that provide valuable assistance to our scientists and researchers. While our rudimentary product is certainly no competition to those from the big players such as Google Scholar, it is clear that with a clear goal and the right set of tools, anyone can build amazing AI-native products for their specific use cases.

