Bota’s Smart Colony-PCR Pipeline: A Synergy of Human and Machines
Background
Here at Bota, we use Colony PCR to screen hundreds of genome transformation designs every week. Colony PCR is a technique used to rapidly screen bacterial colonies for successful gene insertion or knockout after a cloning experiment. While being quicker and less labor-intensive than genome sequencing for initial verification, the traditional Colony PCR method does require some tiring and repetitive manual processes, such as designing optimal PCR primers for each transformation design and reading the PCR results following gel electrophoresis.
We are proud to announce that Bota has designed and built one of the most automated Colony PCR pipelines in the industry, allowing over 500 bacterial transformation designs to be screened every week by just two human operators working 8 hours a day. This pipeline not only improves efficiency and reduces the risk of human error, but also saves our colleagues from boring laborious work and allows them to focus more on making new ideas come true.
Bota’s Solution
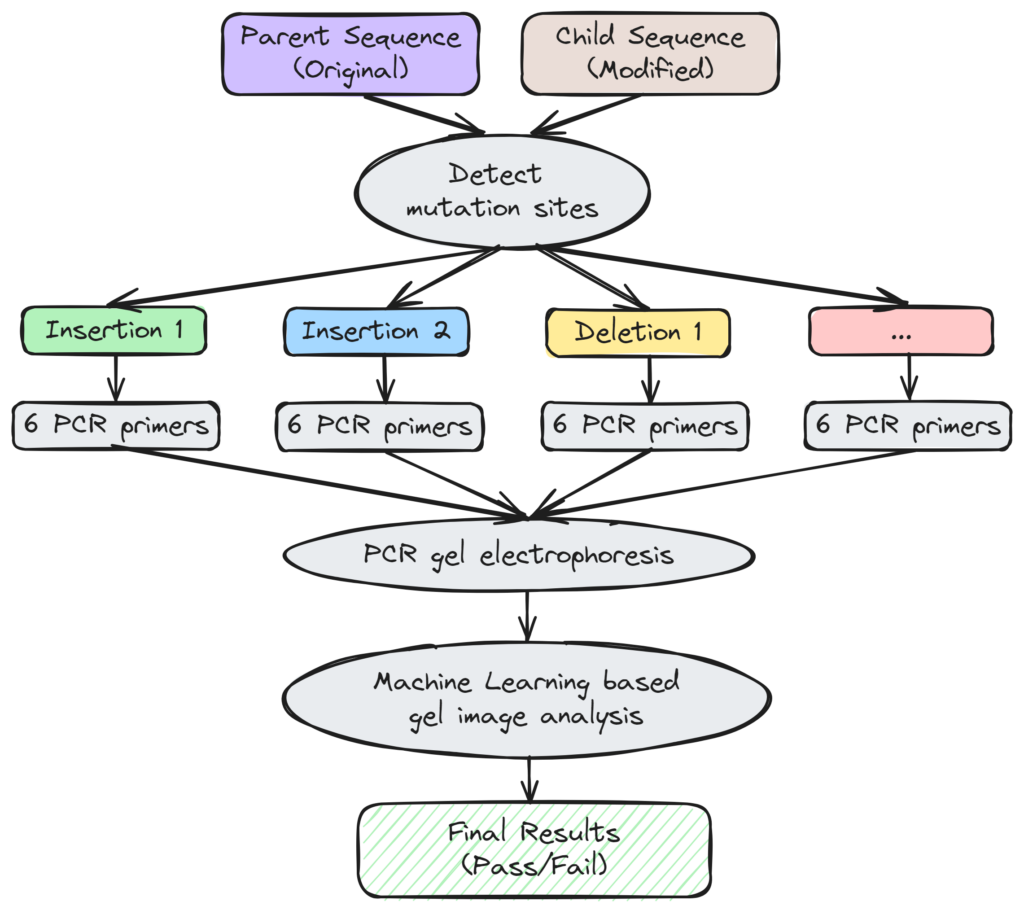
Workflow Diagram
Automatic PCR Primer Design
As the first step to automating the Colony PCR process, we have created a universal algorithm for designing the most optimal PCR primer pairs for any one of the genome transformation designs: insertion (knock-in), removal (knock-out) and promoter swap.
Our universal algorithm is the first in the industry to automatically locate each and every mutation sites (modified regions) by comparing the parent and the child sequence of the transformed colony. The users can finetune several parameters for the mutation site detection step and preview the result within the app, or choose one of our pre-defined parameter sets for common hosts such as the E.coli and Yarrowia. Next, our primer design program will calculate and design 2 primer pairs (4 primers) for both the parent and the child sequence of each mutation site. Since two of the designed primers are supposed to anneal to both the parent and the child sequence, a total of 6 primers will be designed for each and every mutation site.
Smart PCR Gel Image Analyzer
Another time consuming and labor intensive step in a PCR experiment is to read and record the readings from the PCR gel following the gel electrophoresis. With hundreds of designs being processed every week here at Bota, it would take an experienced lab operator hours, if not days to manually inspect and record the readings from thousands of bands from the gels. Not only is this process highly repetitive and time consuming, but also harsh on our operator’s back and eyesight.
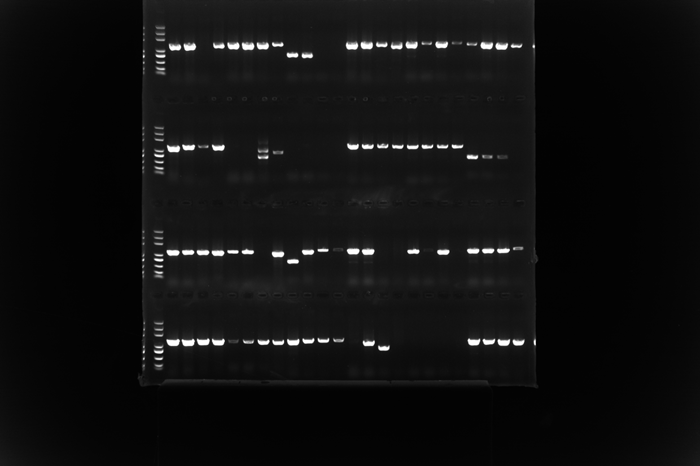
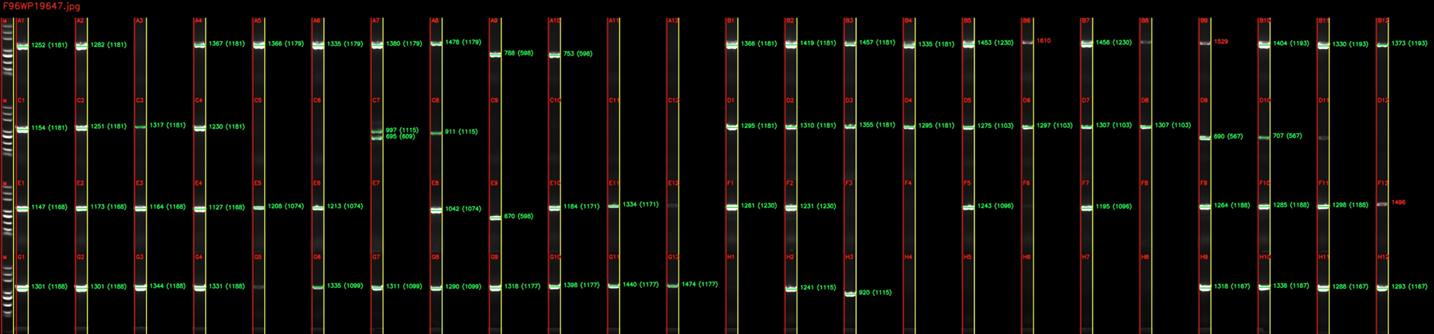
Here comes Bota’s Gel Image Analyzer, our innovative, industry-leading machine learning based program that automatically reads the band sizes from photos of PCR gels. The Gel Image Analyzer uses computer vision to detect each lane from a PCR gel image, corrects any distortion in the gel, and aligns the bands in each lane to the DNA ladder to calculate their sizes. The program then compares the read lengths to their corresponding expected lengths from the PCR primer design program in the previous step, and generates an experiment report containing the pass / fail status of each colonies.
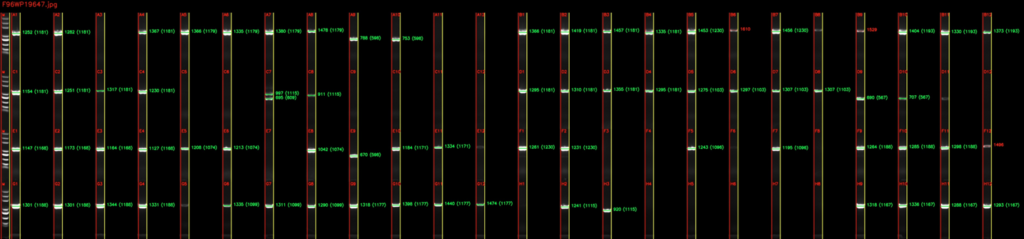
We field tested our Gel Image Analyzer through 3 weeks of actual lab runs, covering over 1500 E.coli colonies. Using experiment records generated by human operators, our Gel Image Analyzer was able to achieve an accuracy rate of over 97%, while only taking minutes instead of days to process batches of gel images.
Summary
The smart colony-PCR pipeline is one of many examples of how we are innovating the use of computational automation in our daily workflow here at Bota. Apart from in-house developed software such as the Automatic PCR Primer Designer and the Gel Image Analyzer, we have also automated physical lab processes such as liquid handling and fermentation sampling through the use of Biomek workstations, robotic arms and auto-samplers. We hope this short article gives you a glimpse of how we are accelerating our R&D processes by working smarter, not harder, through the use of computational biology.
If you have any questions or business inquiries, feel free to reach out to us.

